Software
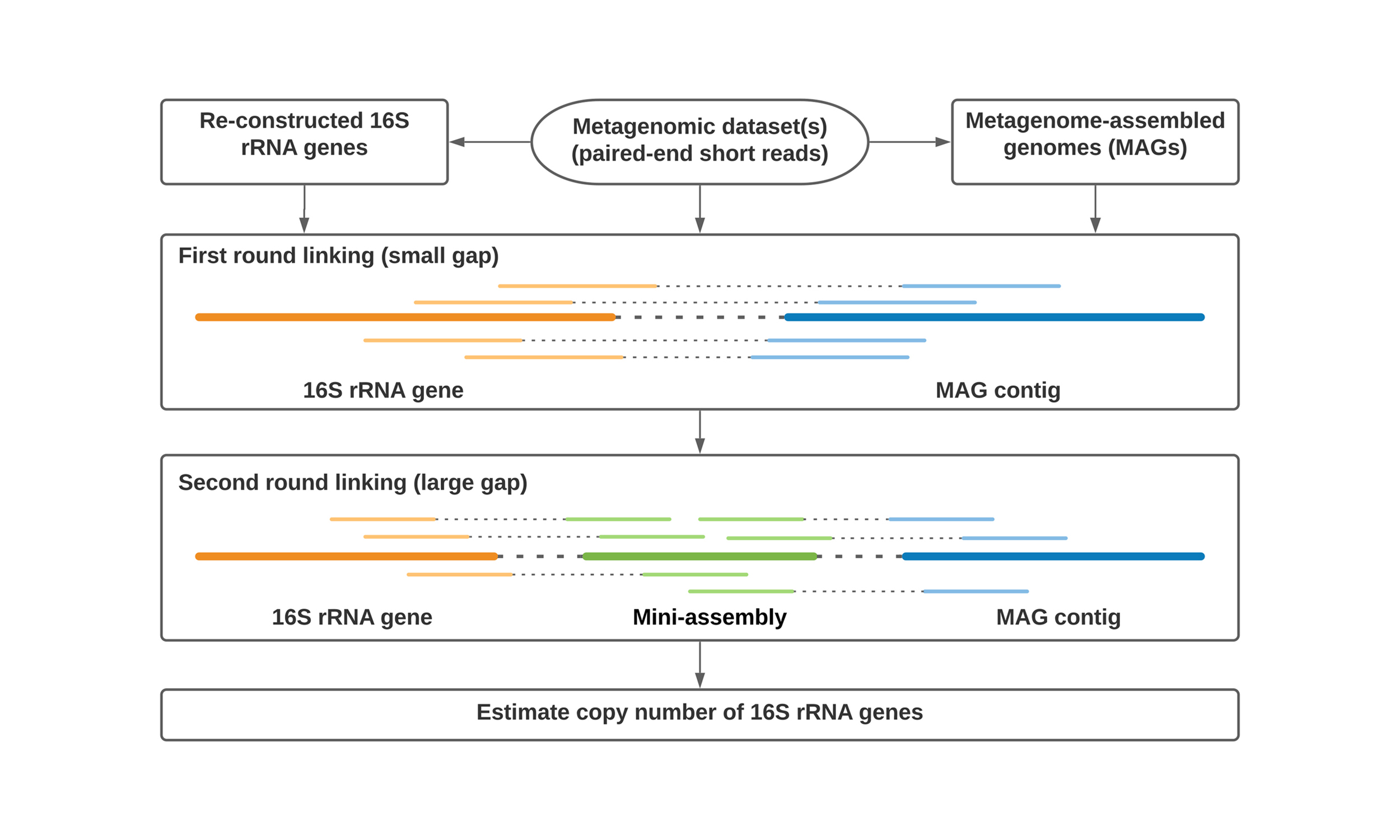
MarkerMAG
Metagenome-assembled genomes (MAGs) have substantially extended our understanding of microbial functionality. However, 16S rRNA genes, which are commonly used in phylogenetic analysis and environmental surveys, are often missing from MAGs. MarkerMAG can link 16S rRNA genes to MAGs using paired-end sequencing reads.
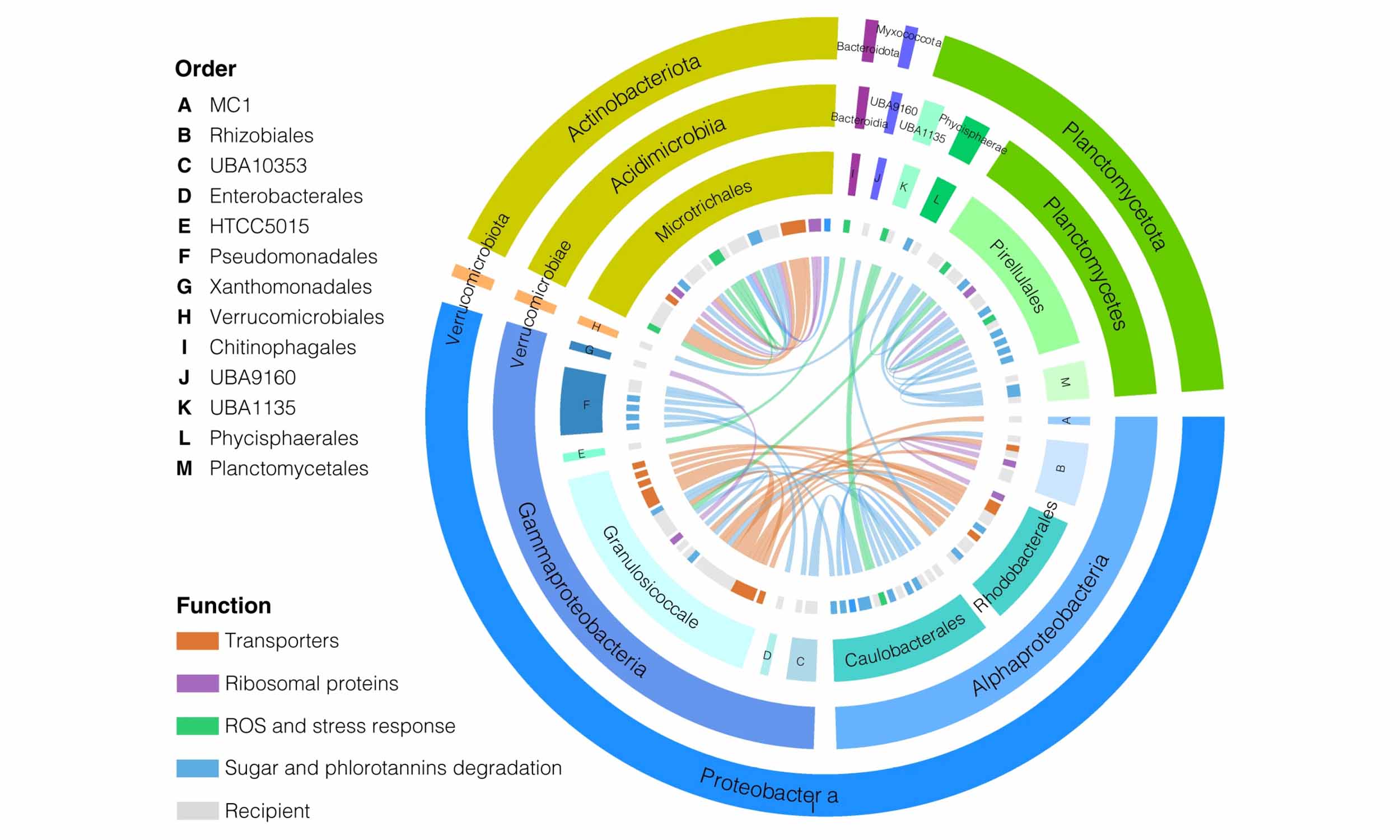
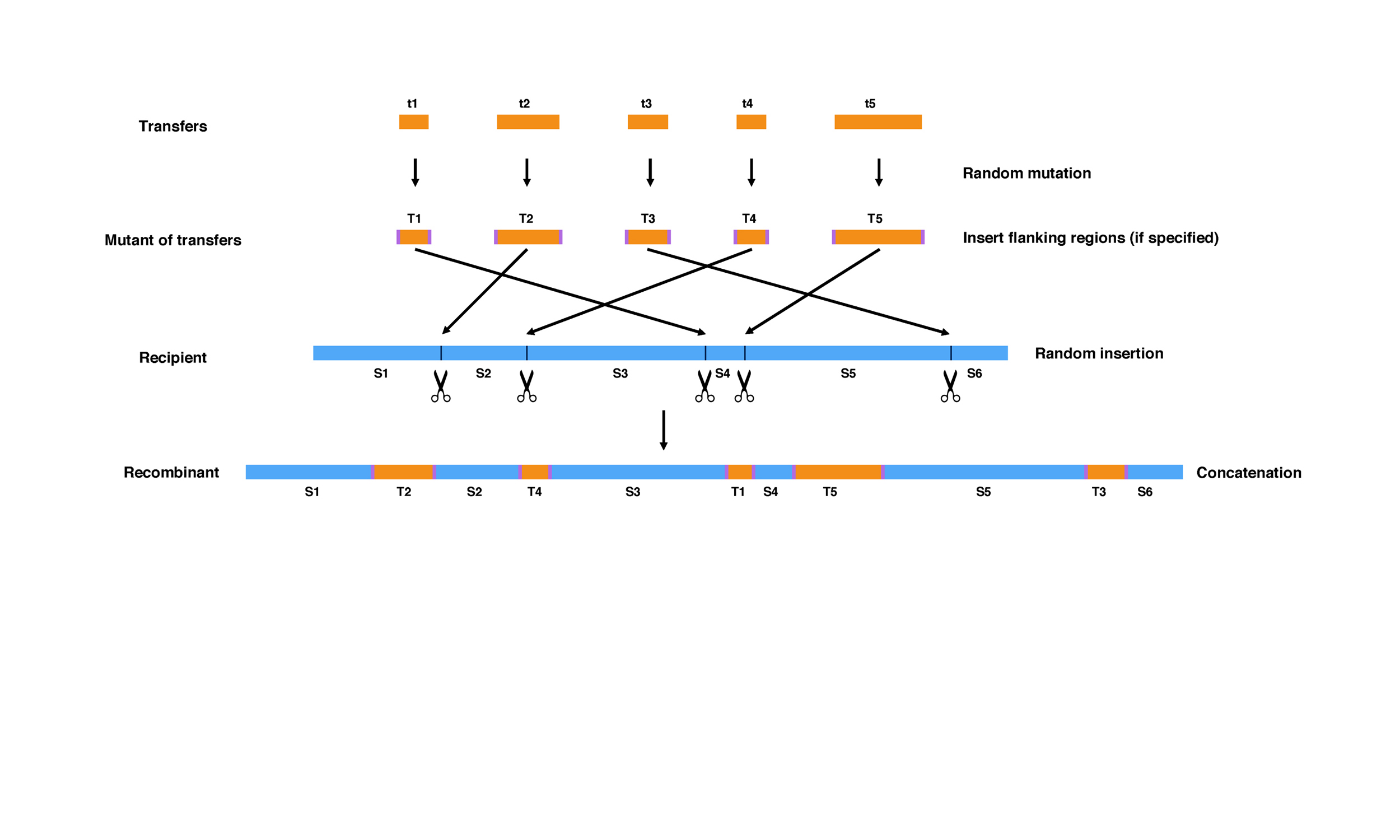
MetaCHIP
Metagenomic datasets provide an opportunity to study horizontal gene transfer (HGT) on the level of a microbial community. However, current HGT detection methods cannot be applied to community-level datasets or require reference genomes. Here, we present MetaCHIP, a pipeline for reference-independent HGT identification at the community level.
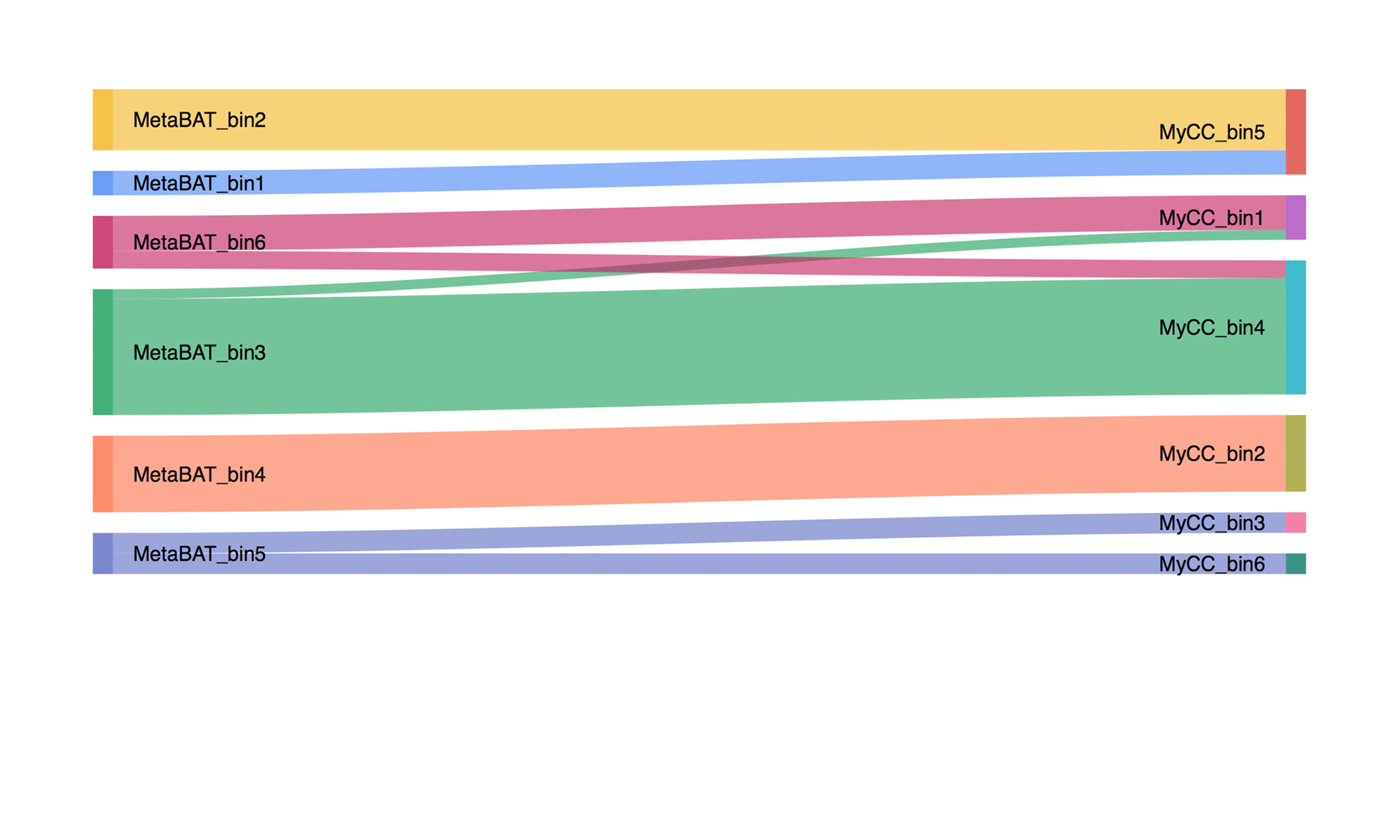
Binning_refiner
Microbial genomes have recently been reconstructed from metagenomic datasets using binning approaches. Inconsistent binning results are however often observed between different binning programs, likely due to the different algorithms or statistical models used. We present Binning_refiner, a pipeline that merges the results of different binning programs.